Mock Lab
MAX-EDER RESEARCH GROUP - Clinical Multi-Omics
MAX-EDER RESEARCH GROUP - Clinical Multi-Omics
© Andreas Mock
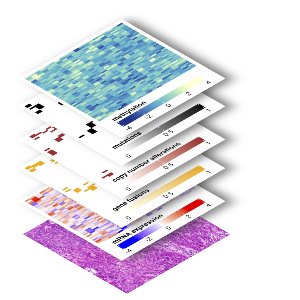
In the fight against cancer, Clinical Multi-Omics is revolutionizing how we understand, diagnose, and treat this complex disease. By integrating diverse biological data - such as epigenomics, genomics, transcriptomics, proteomics, and metabolomics - this approach provides a comprehensive view of tumor biology.
The recent extension with single-cell approaches, spatial profiling, and digital pathology adds unprecedented depth and resolution to cancer research. Single-cell technologies allow for the dissection of tumor heterogeneity by analyzing molecular profiles at the level of individual cells, uncovering rare cell populations and dynamic cellular states. Spatial profiling integrates molecular data with tissue architecture, revealing how tumor cells interact with their microenvironment, including immune and stromal cells. Digital pathology, powered by AI, enables high-throughput, quantitative analysis of tissue samples, linking morphological features to molecular insights.
Together, these innovations provide a more holistic understanding of cancer, paving the way for precise, biology informed diagnostics and therapies.
Handbook - Clinical multi-omics for Precision Oncology - Mock et al. 2023
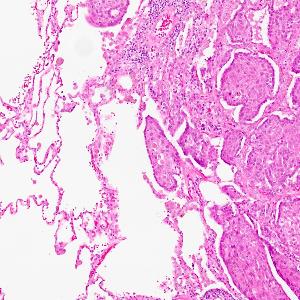
Head and neck cancer cells (right) infiltrate into the lung (left). | © Andreas Mock
Head and neck squamous cell carcinoma (HNSCC) is the seventh most common cancer diagnosis, accounting for approximately 4.5% of cancer-related deaths worldwide. More than 60% of patients present with stage III or IV disease at the time of diagnosis, characterized by large tumors with local invasion and/or metastases to regional lymph nodes. These locally advanced stages of the disease carry a high risk of local recurrence and distant metastasis.
Multimodal approaches (surgery, radiotherapy, and systemic treatments) have improved cure rates, but the prognosis for patients with recurrent disease or distant metastases remains poor.
Using single-cell sequencing, tumor slice cultures, spatial tumor profiling and multimodal integrative data analysis with published data sets, we aim to develop novel therapeutic approaches for incurable recurrent/metastatic head and neck sqamous cell carcinoma.
Our head and neck cancer research is supported by the Max-Eder Junior Research Group Program of the German Cancer Aid (Deutsche Krebshilfe)
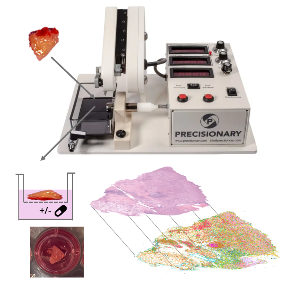
Fresh tumor tissue is obtained from the operating room and processed by a vibrating microtome and cultivated at the air-liquid-interface. Deep biological readouts can be achied by spatial profiling methods. | © Andreas Mock
Organotypic tissue slice cultures provide a groundbreaking platform for functional precision medicine by preserving the native structure, cellular diversity, and microenvironment of patient-derived tissues. These ex vivo models enable personalized drug testing, allowing therapies to be evaluated directly on patient-specific samples for efficacy and resistance. By maintaining interactions between tumor cells, stromal components, and immune cells, they are particularly valuable for studying complex cancer dynamics and immunotherapy responses.
Advances in imaging, spatial profiling, and single-cell analysis are enhancing the learnings from tissue slice cultures, enabling deeper insights into disease mechanisms.
These models bridge the gap between preclinical studies and clinical applications.
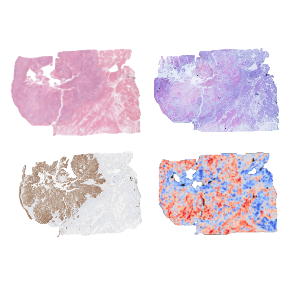
HE staining serve as training data for predictions and performance can be e.g. tested by (multiplex) immunohistochemistry. | © Andreas Mock
Digital pathology integrates high-resolution imaging with computational tools, enabling quantitative analysis and linking tissue morphology to molecular data. AI-powered algorithms detect subtle histological patterns linked to clinical outcomes, enhancing diagnostics and uncovering new biomarkers.
The convergence of these technologies allows for deeper insights into tumor biology, improved patient stratification, and the optimization of targeted therapies. By uniting AI-driven analysis with molecular profiling, this approach accelerates personalized, data-driven cancer care, enabling more precise and effective treatments.
Towards Explainable Artificial Intelligence for Precision Pathology, Klauschen et al. 2024

Assistenzarzt & Arbeitsgruppenleiter
Max-Eder Research Group - Clinical Multi-Omics
Research Associate
Digital Spatial Pathway Mapping Reveals Prognostic Tumor States in Head and Neck Cancer
Hense J, Jamshidi Idaji M, Ciernik L, Dippel J, Ersan F, Knebel M, Pusztai A, Sendelhofert A, Buchstab O, Fröhling S, Otto S, Hess J, Liokatis P, Klauschen F, Müller K-R, Mock A
bioRxiv. 2025 doi: 10.1101/2025.11.24.689710
NCT/DKFZ MASTER handbook of interpreting whole-genome, transcriptome, and methylome data for precision oncology
Mock A*, Teleanu MV*, Kreutzfeldt S, Heilig CE, Hüllein J, Möhrmann L, Jahn A, Hanf D, Kerle IA, Singh HM, Hutter B, Uhrig S, Fröhlich M, Neumann O, Hartig A, Brückmann S, Hirsch S, Grund K, Dikow N, Lipka DB, Renner M, Bhatti IA, Apostolidis L, Schlenk RF, Schaaf CP, Stenzinger A, Schröck E, Hübschmann D, Heining C, Horak P, Glimm H, Fröhling S.
NPJ Precis Oncol. 2023 Oct 26;7(1):109. doi: 10.1038/s41698-023-00458-w.
Transcriptome profiling for precision cancer medicine using shallow nanopore cDNA sequencing
Mock A, Braun M, Scholl C, Fröhling S, Erkut C.
Sci Rep. 2023 Feb 9;13(1):2378. doi: 10.1038/s41598-023-29550-8.
Comprehensive Genomic and Epigenomic Analysis in Cancer of Unknown Primary Guides Molecularly-Informed Therapies Despite Heterogeneity
Möhrmann L*, Werner M*, Oleś M*, Mock A*, Uhrig S, Jahn A, Kreutzfeldt S, Fröhlich M, Hutter B, Paramasivam N, Richter D, Beck K, Winter U, Pfütze K, Heilig CE, Teleanu V, Lipka DB, Zapatka M, Hanf D, List C, Allgäuer M, Penzel R, Rüter G, Jelas I, Hamacher R, Falkenhorst J, Wagner S, Brandts CH, Boerries M, Illert AL, Metzeler KH, Westphalen CB, Desuki A, Kindler T, Folprecht G, Weichert W, Brors B, Stenzinger A, Schröck E, Hübschmann D, Horak P, Heining C, Fröhling S, Glimm H.
Nat Commun. 2022 Aug 2;13(1):4485. doi: 10.1038/s41467-022-31866-4.
Comprehensive Genomic and Transcriptomic Analysis for Guiding Therapeutic Decisions in Patients with Rare Cancers
Horak P* Heining C*, Kreutzfeldt S*, Hutter B*, Mock A**, Hüllein J, Fröhlich M, Uhrig S, Jahn A, Rump A, Gieldon L, Möhrmann L, Hanf D, Teleanu V, Heilig CE, Lipka DB, Allgäuer M, Ruhnke L, Laßmann A, Endris V, Neumann O, Penzel R, Beck K, Richter D, Winter U, Wolf S, Pfütze K, Geörg C, Meißburger B, Buchhalter I, Augustin M, Aulitzky WE, Hohenberger P, Kroiss M, Schirmacher P, Schlenk RF, Keilholz U, Klauschen F, Folprecht G, Bauer S, Siveke JT, Brandts CH, Kindler T, Boerries M, Illert AL, von Bubnoff N, Jost PJ, Spiekermann K, Bitzer M, Schulze-Osthoff K, von Kalle C, Klink B, Brors B, Stenzinger A, Schröck E, Hübschmann D, Weichert W, Glimm H, Fröhling S.
Cancer Discov. 2021 Nov;11(11):2780-2795. doi: 10.1158/2159-8290.CD-21-0126.
Knowledge Bases and Software Support for Variant Interpretation in Precision Oncology
Borchert F*, Mock A*, Tomczak A, Hügel J, Alkarkoukly S, Knurr A, Volckmar AL, Stenzinger A, Schirmacher P, Debus J, Jäger D, Longerich T, Fröhling S, Eils R, Bougatf N, Sax U, Schapranow MP.
Brief Bioinform. 2021 Nov 5;22(6):bbab246. doi: 10.1093/bib/bbab246.
Surfactant Expression Defines an Inflamed Subtype of Lung Adenocarcinoma Brain Metastases that Correlates with Prolonged Survival
Pocha K*, Mock A*, Rapp C, Dettling S, Warta R, Geisenberger C, Jungk C, Martins LR, Grabe N, Reuss D, Debus J, von Deimling A, Abdollahi A, Unterberg A, Herold-Mende CC
Clin Cancer Res. 2020 May 1;26(9):2231-2243. doi: 10.1158/1078-0432.CCR-19-2184.