1. Investigating the dynamics of the neuronal RNP network (funded by the DFG, KI 502/9-1)

The MS2 RNA imaging system consisting of two components (Bertrand et al., 1998; cartoon from: Jambhekar & Derisi, 2007). The GFP-tagged MS2 coat protein (MS2 CP = MCP, plasmid 1) binds the MS2-repeats fused to the target mRNA (plasmid 2) with high affinity, allowing for visualization of individual RNA by live cell imaging in primary neurons.
In this project, we want to study RNP assembly in primary neurons and unravel the underlying dynamics of the RNP network. Therefore, we will exploit a battery of interactor screens complemented with live-cell imaging and state-of-the-art biochemistry to unravel how the RBP network dynamically assembles and balances cellular alterations. In particular, we would like to investigate the following important goals in detail:
- Identification of the RBP protein interactor landscape
- Unravelling the mRNA bound proteome
- Gaining functional insight into RNP assembly dynamics, RNA stability and transport as well as translational control
For this long-term project, two essential experimental approaches are absolutely critical:
1A: the MS2 imaging system to follow the fate of mRNAs in living neurons (established by Karl Bauer in the lab)
We have established an improved version of the MS2 RNA imaging system (Bertrand et al., 1998) with the help of Edouard Bertrand (IGH, Montpellier, France) In brief, the MS2 system utilizes the specific, high affinity interaction between a 19-nucleotide stem-loop structure of the MS2 phage RNA and the MS2 coat protein (MCP) (Figure 1).
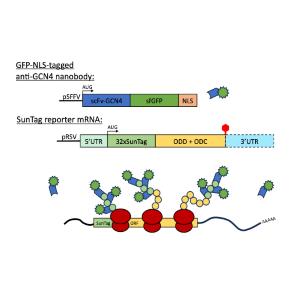
Overview of the SunTag translation imaging system and its use in primary hippocampal neurons. Experimental overview of the SunTag system as used by the Singer and Bertrand laboratories in their recent publication (Pichon et al., 2016, JCB).
Together, this allows us to analyze the assembly and transport dynamics of individual RNP granules in living neurons.
1B: the SunTag imaging system to study local translation in living neurons (established by Karl Bauer in the lab)
Here, we have also established an improved version of the cutting-edge SunTag system (Figure 2), which allows real-time tracking of translation at the single mRNA level (Wu et al., 2016, Science; Pichon et al., 2016, JCB). This system uses a GFP-tagged nanobody that binds to a 32-repeat peptide derived from yeast GCN4, which is encoded in the 5’-end of the nascent transcript. When translation is initiated, ribosomes synthesize the peptide repeats, which are then recognized by the GFP-tagged nanobody, leading to the visible clustering of GFP at translation sites. As polysomes form, GFP clustering intensifies, reaching maximum brightness and allowing precise monitoring of active translation until termination, when the signal disperses. The SunTag system is established in the Kiebler lab in primary neuron cell culture (collaboration with Prof. Edouard Bertrand, Montpellier, F).
Together, this allows us to track translation dynamics in real-time through live microscopy in neurons.
